Data Density Plots¶
This is a comparison to the Exploring normalizations Matplotlib example.
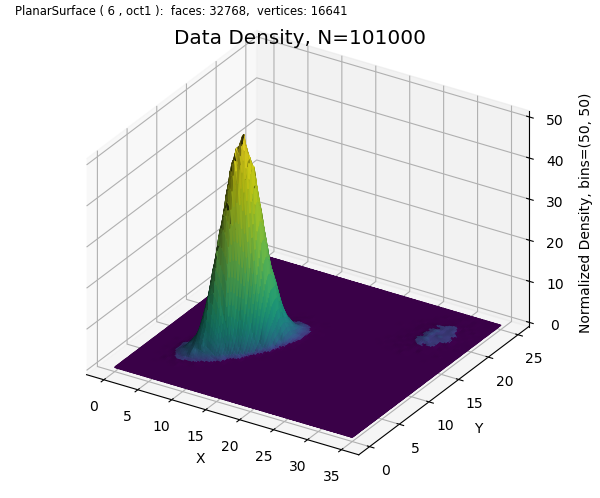
This example demonstrates the use of the S3Dlib functions:
density_function returns a function, f(x,y), which is created from a set of x,y coordinates argument and a bins argument analogous to 2D histogram creation methods. The returned function,f, computes the data density at x,y coordinates passed to the function. The volume of density surface is normalized to 1 but may be scaled using the scale argument.
op_cmap is a S3Dlib colormap utility function which generates a colormap from a function which is passed as an argument. In this example, the function passed, power, provided a colormap equivalent to the Matplotlib colors.PowerNorm
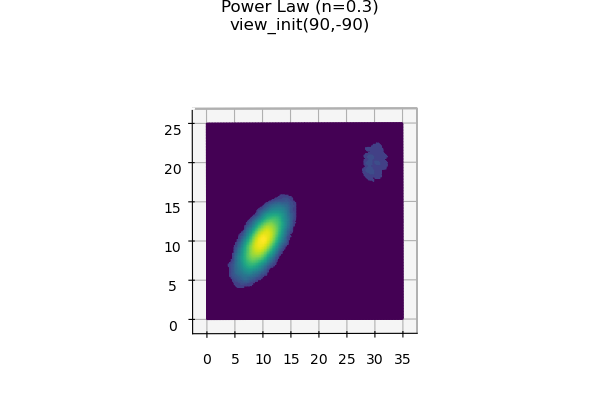
The 2D plot can be visualized by rotating the surface using ax.view_init(90,-90). The angles used for view_init were offset from 90 to force axis label positions in the 3D rendering. The result is a figure similar to a 2D histogram as shown below.
import numpy as np
from numpy.random import multivariate_normal
import matplotlib.pyplot as plt
from matplotlib import cm,colormaps
import s3dlib.surface as s3d
import s3dlib.cmap_utilities as cmu
#.. Matplotlib Examples: 3D surface histogram with normalization distribution
# 1. Define function to examine .....................................
rez, bns, N = 6, (50,50), 150
np.random.seed(19680801)
data = np.vstack([
multivariate_normal([10, 10], [[3, 2], [2, 3]], size=100000),
multivariate_normal([30, 20], [[3, 1], [1, 3]], size=1000)
])
data = data.T
f = s3d.density_function(data,bins=bns,scale=True)
def surfDist(xyz):
x,y,z=xyz
return x,y,f(x,y) # use density function for Z
def power(t,n) :
return colormaps['viridis'](np.power(t,n)).T
cmap= cmu.op_cmap(lambda t: power(t,0.3), name='Power Law (n=0.3)')
# FIGURE 1 : Density Surface ====================================================
# ===============================================================================
# 2. Setup and map surfaces .........................................
surface = s3d.PlanarSurface(rez,'oct1').domain([0,35],[0,25])
surface.map_geom_from_op(surfDist)
surface.map_cmap_from_op(cmap=cmap)
# 3. Construct figure, add surface, plot ............................
fig = plt.figure(figsize=(6,5))
fig.text(0.025,0.99,str(surface), ha='left', va='top', fontsize='smaller')
ax = plt.axes(projection='3d')
s3d.auto_scale(ax,surface)
ax.add_collection3d(surface.shade())
zlabel = 'Normalized Density, bins='+str(bns)
ax.set(xlabel='X',ylabel='Y',zlabel=zlabel)
ax.set_proj_type('ortho')
ax.view_init(35,-58)
title = "Data Density, N="+ str(len(data[0]))
ax.set_title(title , fontsize='x-large' )
fig.tight_layout()
# FIGURE 2 : Top View of Density Surface ========================================
# ===============================================================================
surface2 = s3d.PlanarSurface(rez,'oct1').domain([0,35],[0,25])
surface2.map_geom_from_op(surfDist)
surface2.map_cmap_from_op(cmap=cmap)
# 3. Construct figure, add surface, plot ............................
fig = plt.figure(figsize=(6,4))
ax2 = plt.axes(projection='3d')
s3d.auto_scale(ax2,surface2)
ax2.add_collection3d(surface2)
ax2.set(xlabel='',ylabel='')
ax2.tick_params("z", colors='w')
ax2.set_proj_type('ortho')
ax2.view_init(89.7,-90.1)
ax2.set_title( cmap.name+'\nview_init(90,-90)\n', va='top' )
Data points can be individually assigned color using the density function, f, as shown below.
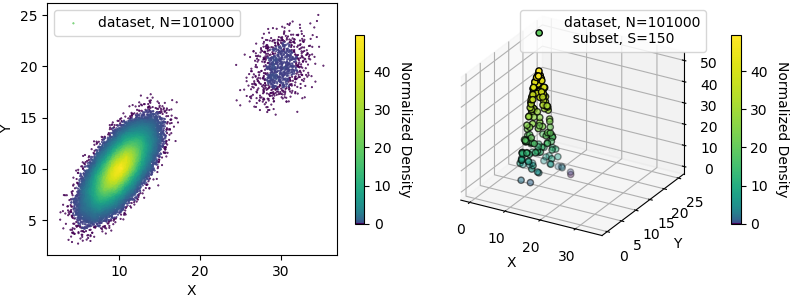
# FIGURE 3 : 2d & 3D plots of data set and sub-set ==============================
# ===============================================================================
vals = f(*data)
fvals = vals/np.amax(vals) # normalize to range [0,1]
colors = [ cmap(x) for x in fvals ]
subset_data = data[:,:N]
subset_colors = colors[:N]
x,y,z = subset_data[0], subset_data[1], f(*subset_data) # use density function for Z
label_1 = "dataset, N="+str(len(data[0]))
label_2 = label_1 + "\n subset, S="+str(len(x))
fig = plt.figure(figsize=(8,3.0))
ax1 = fig.add_subplot(121)
ax2 = fig.add_subplot(122, projection='3d')
ax1.set( xlabel='X', ylabel='Y')
ax2.set( xlabel='X', ylabel='Y')
cbar1 = plt.colorbar(surface.cBar_ScalarMappable, ax=ax1, shrink=0.75 )
cbar1.set_label('Normalized Density', rotation=270, labelpad = 15)
cbar2 = plt.colorbar(surface.cBar_ScalarMappable, ax=ax2, shrink=0.75 )
cbar2.set_label('Normalized Density', rotation=270, labelpad = 15)
ax1.scatter(*data,s=1,marker='.',c=colors,label=label_1)
ax1.legend()
ax2.set_proj_type('ortho')
ax2.scatter(x,y,z,edgecolor='k',c=subset_colors,label=label_2)
ax2.legend()
s3d.auto_scale(ax2,surface)
fig.tight_layout(pad=0)
# ==============================================================================
plt.show()
